Extracting researchers based on affiliations and publications history¶
The purpose of this notebook is to demonstrate how to extract researchers data using the Dimensions API.
Specifically, we want to find out about all researchers based on these two criteria:
Affiliation. That is, whether they are / have been affiliated to a specific GRID organization
Publications. Whether they have published within a chosen time frame.
[2]:
import datetime
print("==\nCHANGELOG\nThis notebook was last run on %s\n==" % datetime.date.today().strftime('%b %d, %Y'))
==
CHANGELOG
This notebook was last run on Jan 25, 2022
==
Prerequisites¶
This notebook assumes you have installed the Dimcli library and are familiar with the ‘Getting Started’ tutorial.
[3]:
!pip install dimcli -U --quiet
import dimcli
from dimcli.utils import *
import os, sys, time, json
import pandas as pd
from IPython.display import Image
from IPython.core.display import HTML
print("==\nLogging in..")
# https://digital-science.github.io/dimcli/getting-started.html#authentication
ENDPOINT = "https://app.dimensions.ai"
if 'google.colab' in sys.modules:
import getpass
KEY = getpass.getpass(prompt='API Key: ')
dimcli.login(key=KEY, endpoint=ENDPOINT)
else:
KEY = ""
dimcli.login(key=KEY, endpoint=ENDPOINT)
dsl = dimcli.Dsl()
Searching config file credentials for 'https://app.dimensions.ai' endpoint..
==
Logging in..
Dimcli - Dimensions API Client (v0.9.6)
Connected to: <https://app.dimensions.ai/api/dsl> - DSL v2.0
Method: dsl.ini file
Select an organization ID¶
For the purpose of this exercise, we will are going to use grid.258806.1 and the time frame 2013-2018. Feel free though to change the parameters below as you want, eg by choosing another GRID organization.
[4]:
# sample org: grid.258806.1
GRIDID = "grid.258806.1" #@param {type:"string"}
START_YEAR = 2013 #@param {type:"slider", min:1900, max:2020, step: 1}
END_YEAR = 2018 #@param {type:"slider", min:1900, max:2020, step: 1}
if START_YEAR < END_YEAR: START_YEAR = END_YEAR
YEARS = f"[{START_YEAR}:{END_YEAR}]"
Background: understanding the data model¶
In order to process researchers affiliation data in the context of publications, we should first take the time to understand how this data is structured in Dimensions.
The JSON results of any query with shape search publications where .... return publications are composed by a list of publications. If we open up one single publication record we will immediately see that authors are stored in a nested object authors that contains a list of dictionaries. Each element in this dictionary represents one single publication author and includes other information e.g. name, surname, ID, the organizations he/she is affiliated with etc..
For example, in order to extract the second author of the tenth publication from our results we would do the following: results.publications[10]['authors'][1]:
# author info
...
{'first_name': 'Noboru',
'last_name': 'Sebe',
'orcid': '',
'current_organization_id': 'grid.258806.1',
'researcher_id': 'ur.010647607673.28',
'affiliations': [{'id': 'grid.258806.1',
'name': 'Kyushu Institute of Technology',
'city': 'Kitakyushu',
'city_id': 1859307,
'country': 'Japan',
'country_code': 'JP',
'state': None,
'state_code': None}]}
...
Here’s a object model diagram summing up how data is structured.
[5]:
Image(url= "http://api-sample-data.dimensions.ai/diagrams/data_model_researchers_publications1.v2.jpg", width=800)
[5]:
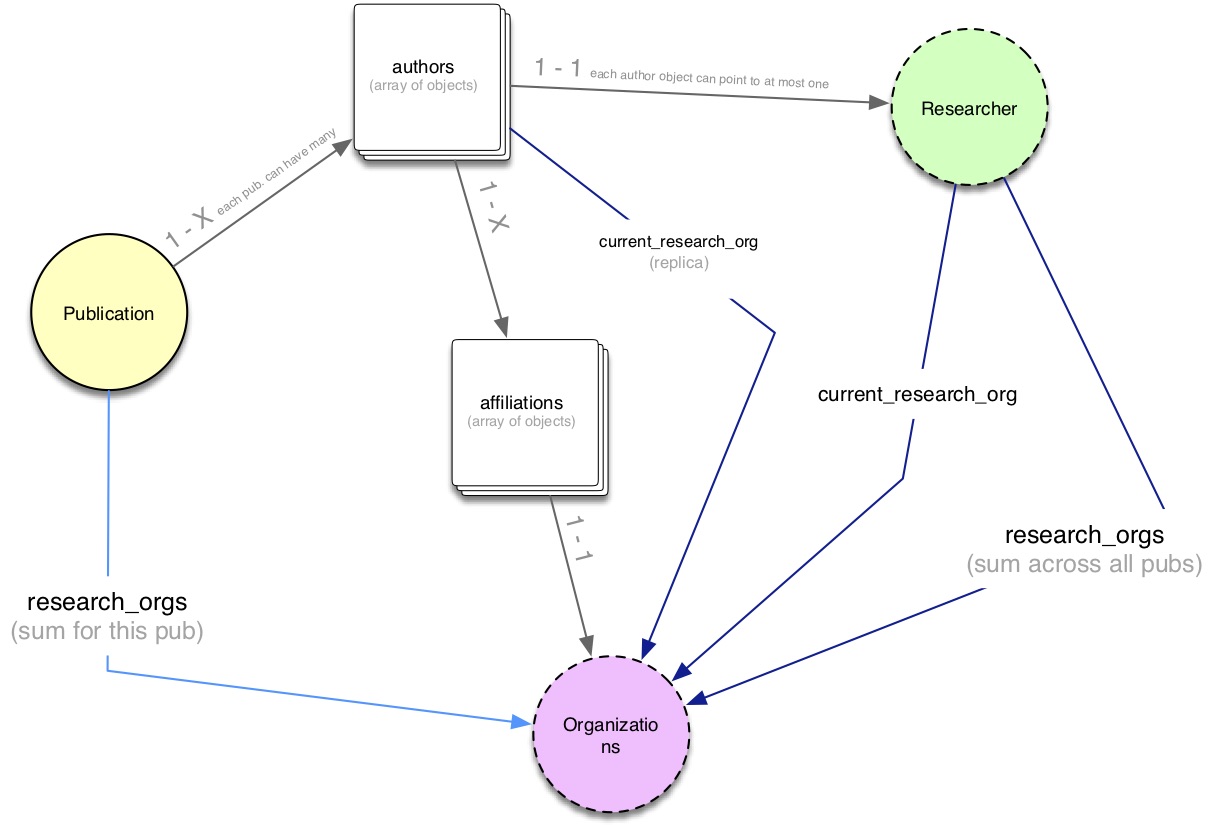
There are a few important things to keep in mind:
Publication Authors VS Researchers. In Dimensions, publication authors don’t necessarily have a
researcherID (eg because they haven’t been disambiguated yet). So a publication may have N authors (stored in JSON within theauthorskey), but only a subset of these include aresearcher_idlink. PS see also the searching for researchers section of the API docs for more info on this topic.Time of the affiliation. Researchers can be affiliated to a specific GRID organization either at the time of speaking (now), or at the time of writing (i.e. when the article was published). The DSL uses different properties to express this fact:
current_research_orgor simplyresearch_orgs.Denormalized fields. Both the Publication and Researcher sources include a
research_orgsfield. In both cases that’s a denormalized field ie a ‘shortcut version’ of data primarily found in theauthorsfield of publications. However, although they share the same name, the tworesearch_orgsfields don’t have the same meaning.In publications,
research_orgscontains the set of all authors’ affiliations for a single article.In researchers,
research_orgscontains the set of all research organizations a single individual has been affiliated to throughout his/her career (as far as Dimensions knows, of course!).
So, in the real world we often have scenarios like the following one:
[6]:
Image(url= "http://api-sample-data.dimensions.ai/diagrams/data_model_researchers_publications2.v2.jpg", width=1000)
[6]:
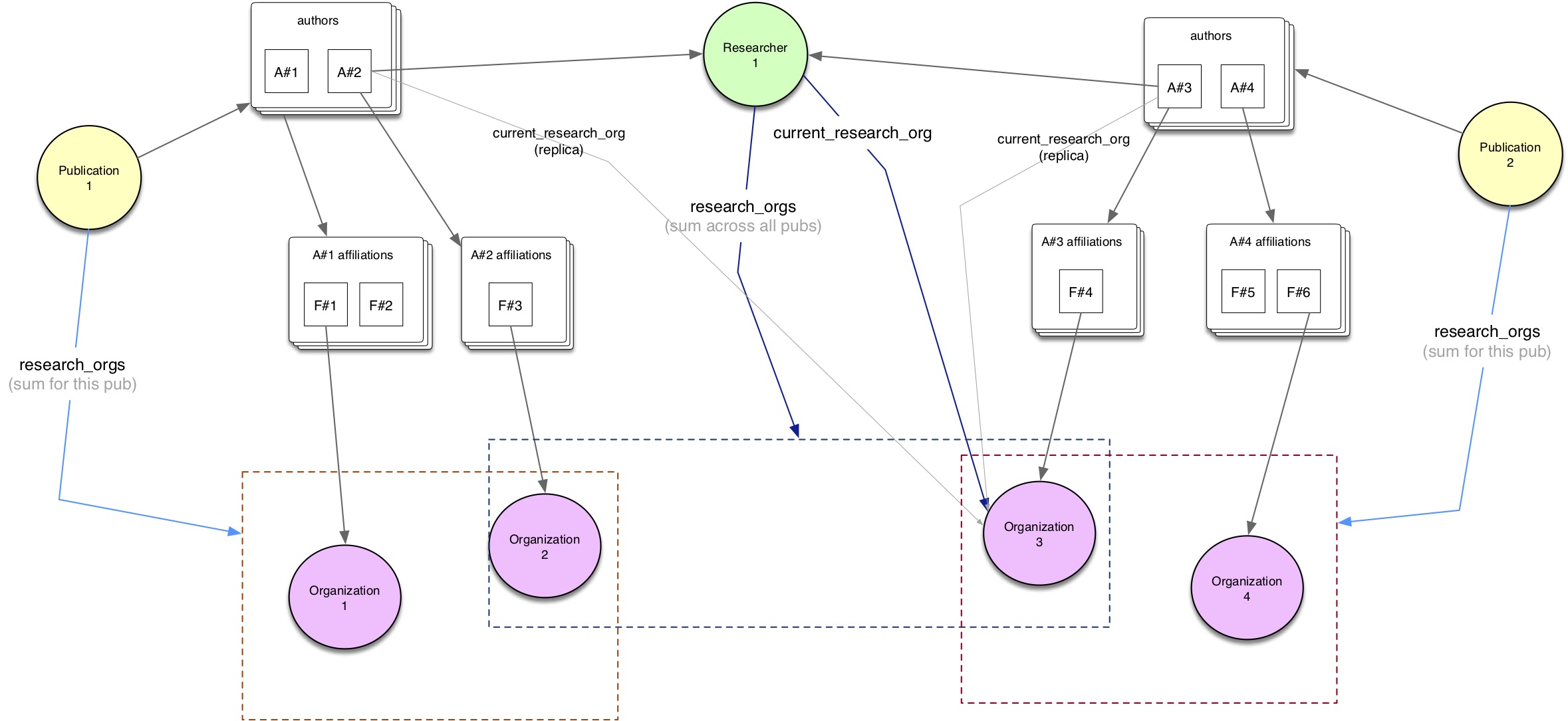
Methodology: two options available¶
It turns out that there are two possible ways to extract these data, depending on whether we start our queries from Publications or from Researchers.
Starting from the Publication source, we can first filter publications based on our constraints (eg year range
[2013-2018]andresearch_orgs="grid.258806.1"- but it could be any other query parameters); second, we would loop over all of these publications so to extract all relevant reasearchers using theaffiliationdata.Starting from the Researcher source, we would first filter researchers based on our constraints (eg with
research_orgs="grid.258806.1"); second, we would search for publications linked to these researchers, which have been published in the time frame[2013-2018]; lastly, we extract all relevant reasearchers using theaffiliationdata.
As we will find out later, both approaches are perfectly valid and return the same results.
The first apporach is generally quicker as it has only two steps (while the second one has three).
In real-world situations though, deciding which approach is best depends on the specific query filters being used and on the impact these filters have on the overall performance/speed of the data extraction. There is no fixed rule and a bit of trial & error can go a long way in helping you optimize your data extraction algorithm!
Approach 1. From publications to researchers¶
Starting from the publications source, the steps are as follows:
Filtering publications for year range [2013-2018] and research_orgs=“grid.258806.1”
i.e.
search publications where year in [2013:2018] and research_orgs="grid.258806.1" return publications
Looping over publications’ authors and extracting relevant researchers
if
['affiliation']['id'] == "grid.258806.1"=> that gives us the affiliations at the time of publishing
if
['current_organization_id'] == "grid.258806.1"=> that gives us the affiliations at the time of speaking
First off, we can to get all publications based on our search criteria by using a loop query.
[7]:
pubs = dsl.query_iterative(f"""search publications
where year in {YEARS} and research_orgs="{GRIDID}"
return publications[id+title+doi+year+type+authors+journal+issue+volume]""")
Starting iteration with limit=1000 skip=0 ...
0-1000 / 1052 (1.94s)
1000-1052 / 1052 (1.28s)
===
Records extracted: 1052
First we want to know how many researchers linked to these publications were affiliated to the GRID organization when the publication was created (note: they may be at at a different institution right now).
The affiliation data in publications represent exactly that: we can thus loop over them (for each publication/researcher) and keep only the ones matching our GRID ID.
TIP affiliations can be extracted easily thanks one of the ‘dataframe’ transformation methods in Dimcli: as_dataframe_authors_affiliations
[8]:
# extract affiliations from a publications list
affiliations = pubs.as_dataframe_authors_affiliations()
# select only affiliations for GRIDID
authors_historical = affiliations[affiliations['aff_id'] == GRIDID].copy()
# remove duplicates by eliminating publication-specific data
authors_historical.drop(columns=['pub_id'], inplace=True)
authors_historical.drop_duplicates('researcher_id', inplace=True)
print(f"===\nResearchers with affiliation to {GRIDID} at time of writing:", authors_historical.researcher_id.nunique(), "\n===")
# preview the data
authors_historical
===
Researchers with affiliation to grid.258806.1 at time of writing: 830
===
[8]:
| aff_city | aff_city_id | aff_country | aff_country_code | aff_id | aff_name | aff_raw_affiliation | aff_state | aff_state_code | researcher_id | first_name | last_name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Department of Electrical and Electronic Engine... | Meng | Ge | |||
| 5 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Department of Control Engineering, Kyushu Inst... | ur.0724144151.56 | Joo Kooi | Tan | ||
| 6 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Department of Electrical Engineering, Kyushu I... | ur.010214262704.60 | Gaber | Magdy | ||
| 11 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Department of Electrical Engineering, Kyushu I... | ur.011456332771.53 | Yasunori | Mitani | ||
| 14 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Kyushu Institute of Technology, Kitakyushu, 80... | ur.013152454441.15 | Kazuhiro | Toyoda | ||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5278 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | 九州工業大学 | ur.013230043761.21 | 幸左 | 賢二 | ||
| 5283 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Kyushu Institute of Technology | ur.015230506045.34 | Hirohisa | ISOGAI | ||
| 5290 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Kyushu Institute of Technology, Japan | ur.012270025364.95 | Naoya | Higuchi | ||
| 5291 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Kyushu Institute of Technology, Japan | ur.013003725541.53 | Yasunobu | Imamura | ||
| 5294 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Kyushu Institute of Technology, Japan | ur.015310517641.55 | Takeshi | Shinohara |
830 rows × 12 columns
Note: * The first ‘publication-affiliations’ dataframe we get may contain duplicate records - eg if an author has more than one publication it’ll be listed twice. That’s why we have an extra step where we drop the pub_id column and simply count unique researchers, based on their researcher ID.
This can be achieved simply by taking into consideration a different field, called current_organization_id, available at the outer level of the JSON author structure (see the data model section above) - outside the affiliations list.
Luckily Dimcli includes another handy method for umpacking authors into a dataframe: as_dataframe_authors
[9]:
authors_current = pubs.as_dataframe_authors()
authors_current = authors_current[authors_current['current_organization_id'] == GRIDID].copy()
authors_current.drop(columns=['pub_id'], inplace=True)
authors_current.drop_duplicates('researcher_id', inplace=True)
print(f"===\nResearchers with affiliation to {GRIDID} at the time of speaking:", authors_current.researcher_id.nunique(), "\n===")
authors_current
===
Researchers with affiliation to grid.258806.1 at the time of speaking: 584
===
[9]:
| affiliations | corresponding | current_organization_id | first_name | last_name | orcid | raw_affiliation | researcher_id | |
|---|---|---|---|---|---|---|---|---|
| 5 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Joo Kooi | Tan | None | [Department of Control Engineering, Kyushu Ins... | ur.0724144151.56 | |
| 9 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Yasunori | Mitani | None | [Department of Electrical Engineering, Kyushu ... | ur.011456332771.53 | |
| 12 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Kazuhiro | Toyoda | None | [Kyushu Institute of Technology, Kitakyushu, 8... | ur.013152454441.15 | |
| 13 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Mengu | Cho | None | [Kyushu Institute of Technology, Kitakyushu, 8... | ur.013152112405.28 | |
| 14 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Siowhwa | Teo | [0000-0001-9927-8309] | [Graduate School of Life Science and Systems E... | ur.010525770330.49 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4763 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | 古川 | 徹生 | None | [九州工業大学大学院生命体工学研究科] | ur.013546017565.95 | |
| 4770 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | 西田 | 治男 | None | [九州工業大学大学院生命体工学研究科] | ur.07662175025.31 | |
| 4784 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Naoya | Higuchi | None | [Kyushu Institute of Technology, Japan] | ur.012270025364.95 | |
| 4785 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Yasunobu | Imamura | None | [Kyushu Institute of Technology, Japan] | ur.013003725541.53 | |
| 4788 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Takeshi | Shinohara | None | [Kyushu Institute of Technology, Japan] | ur.015310517641.55 |
584 rows × 8 columns
Approach 2. From researchers to publications¶
Using this approach, we start our search from the ‘researchers’ database (instead of the ‘publications’ database).
There are 3 main steps:
Filtering researchers with research_orgs=GRID-ID (note: this gives us affiliated researches at any point in time)
search researchers where research_orgs="grid.258806.1" return researchers
Searching for publications linked to these researchers and linked to GRID-ID, which have been published in the time frame
[2013-2018]search publications where researchers.id in {LIST OF IDS} and year in [2013:2018] and research_orgs="grid.258806.1" return publicationsNOTE: this a variation of the Approach-1 query above: we have just added the researchers IDs filter (thus reducing the search space)
Extracting relevant researchers from publications, using the same exact steps as in approach 1 above.
if
['current_organization_id'] == "grid.258806.1"=> that gives us the affiliations at the time of speaking
or if
['affiliation']['id'] == "grid.258806.1"=> that gives us the affiliations at the time of publishing
[10]:
q = f"""search researchers where research_orgs="{GRIDID}"
return researchers[basics]"""
researchers_json = dsl.query_iterative(q)
researchers = researchers_json.as_dataframe()
researchers.head()
Starting iteration with limit=1000 skip=0 ...
0-1000 / 11838 (5.43s)
1000-2000 / 11838 (3.73s)
2000-3000 / 11838 (2.46s)
3000-4000 / 11838 (1.84s)
4000-5000 / 11838 (3.44s)
5000-6000 / 11838 (2.64s)
6000-7000 / 11838 (2.29s)
7000-8000 / 11838 (1.98s)
8000-9000 / 11838 (2.25s)
9000-10000 / 11838 (2.40s)
10000-11000 / 11838 (3.11s)
11000-11838 / 11838 (3.06s)
===
Records extracted: 11838
[10]:
| first_name | id | last_name | research_orgs | orcid_id | |
|---|---|---|---|---|---|
| 0 | Makoto | ur.07777767140.16 | Kobayashi | [{'acronym': 'KIT', 'city_name': 'Kitakyushu',... | NaN |
| 1 | Ashraful M | ur.07777460411.31 | Hoque | [{'city_name': 'Houston', 'country_name': 'Uni... | [0000-0002-3970-8538] |
| 2 | Fujio | ur.07777374057.66 | Kubo | [{'city_name': 'Hiroshima', 'country_name': 'J... | NaN |
| 3 | Muhammad Akmal Kamarudin | ur.07777204200.43 | Kamarudin | [{'acronym': 'UM', 'city_name': 'Kuala Lumpur'... | [0000-0002-2256-5948] |
| 4 | Chinatsu | ur.0777713743.09 | Ikeura | [{'acronym': 'KIT', 'city_name': 'Kitakyushu',... | NaN |
Now we need to select only the researchers who have published in the time frame [2013:2018]. So for each researcher ID we must extract the full publication history in order to verify their relevance.
The most efficient way to do this is to use a query that extracts the publication history for several researchers at the same time (so to avoid overruning our API quota), then, as a second step, producing a clean list of relevant researchers from it.
[15]:
results = []
researchers_ids = list(researchers['id'])
# no of researchers IDs per query: so to ensure we never hit the 1000 records limit per query
CHUNKS_SIZE = 300
q = """search publications
where researchers.id in {}
and year in {}
and research_orgs="{}"
return publications[id+title+doi+year+type+authors+journal+issue+volume] limit 1000"""
for chunk in chunks_of(researchers_ids, size=CHUNKS_SIZE):
data = dsl.query(q.format(json.dumps(chunk), YEARS, GRIDID))
try:
results += data.publications
except:
pass
print("---\nFound", len(results), "publications for the given criteria (including duplicates)")
# simulate a DSL payload using Dimcli
pubs_v2 = dimcli.DslDataset.from_publications_list(results)
# transform to a dataframe to remove duplicates quickly
pubs_v2_df = pubs_v2.as_dataframe()
pubs_v2_df.drop_duplicates("id", inplace=True)
print("Final result:", len(pubs_v2_df), "unique publications")
Returned Publications: 59 (total = 59)
Time: 1.99s
Returned Publications: 49 (total = 49)
Time: 0.93s
Returned Publications: 48 (total = 48)
Time: 0.93s
Returned Publications: 61 (total = 61)
Time: 1.02s
Returned Publications: 36 (total = 36)
Time: 0.86s
Returned Publications: 41 (total = 41)
Time: 0.85s
Returned Publications: 64 (total = 64)
Time: 0.94s
Returned Publications: 36 (total = 36)
Time: 0.91s
Returned Publications: 29 (total = 29)
Time: 0.98s
Returned Publications: 48 (total = 48)
Time: 0.93s
Returned Publications: 40 (total = 40)
Time: 0.90s
Returned Publications: 23 (total = 23)
Time: 0.79s
Returned Publications: 45 (total = 45)
Time: 0.93s
Returned Publications: 53 (total = 53)
Time: 1.13s
Returned Publications: 38 (total = 38)
Time: 0.94s
Returned Publications: 61 (total = 61)
Time: 0.96s
Returned Publications: 36 (total = 36)
Time: 1.76s
Returned Publications: 59 (total = 59)
Time: 1.06s
Returned Publications: 52 (total = 52)
Time: 0.94s
Returned Publications: 47 (total = 47)
Time: 0.96s
Returned Publications: 25 (total = 25)
Time: 0.88s
Returned Publications: 52 (total = 52)
Time: 0.98s
Returned Publications: 67 (total = 67)
Time: 0.98s
Returned Publications: 72 (total = 72)
Time: 1.02s
Returned Publications: 26 (total = 26)
Time: 0.83s
Returned Publications: 64 (total = 64)
Time: 0.95s
Returned Publications: 52 (total = 52)
Time: 0.96s
Returned Publications: 42 (total = 42)
Time: 0.85s
Returned Publications: 75 (total = 75)
Time: 0.94s
Returned Publications: 81 (total = 81)
Time: 1.01s
Returned Publications: 99 (total = 99)
Time: 1.06s
Returned Publications: 22 (total = 22)
Time: 0.91s
Returned Publications: 109 (total = 109)
Time: 0.94s
Returned Publications: 48 (total = 48)
Time: 0.91s
Returned Publications: 38 (total = 38)
Time: 0.85s
Returned Publications: 78 (total = 78)
Time: 0.92s
Returned Publications: 53 (total = 53)
Time: 0.89s
Returned Publications: 72 (total = 72)
Time: 1.03s
Returned Publications: 50 (total = 50)
Time: 1.06s
Returned Publications: 15 (total = 15)
Time: 0.69s
---
Found 2065 publications for the given criteria (including duplicates)
Final result: 904 unique publications
This step is basically the same as in approach 1 above.
[16]:
# extract affiliations from a publications list
affiliations_v2 = pubs_v2.as_dataframe_authors_affiliations()
# select only affiliations for GRIDID
authors_historical_v2 = affiliations_v2[affiliations_v2['aff_id'] == GRIDID].copy()
# remove duplicates by eliminating publication-specific data
authors_historical_v2.drop(columns=['pub_id'], inplace=True)
authors_historical_v2.drop_duplicates('researcher_id', inplace=True)
print(f"===\nResearchers with affiliation to {GRIDID} at time of writing:", authors_historical_v2.researcher_id.nunique(), "\n===")
# preview the data
authors_historical_v2
===
Researchers with affiliation to grid.258806.1 at time of writing: 740
===
[16]:
| aff_city | aff_city_id | aff_country | aff_country_code | aff_id | aff_name | aff_raw_affiliation | aff_state | aff_state_code | researcher_id | first_name | last_name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Graduate School of Life Science and Systems En... | ur.010525770330.49 | Siowhwa | Teo | ||
| 1 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Graduate School of Life Science and Systems En... | ur.07706074430.00 | Zhanglin | Guo | ||
| 2 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Graduate School of Life Science and Systems En... | ur.012076416030.50 | Zhenhua | Xu | ||
| 3 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Graduate School of Life Science and Systems En... | ur.010227671107.36 | Chu | Zhang | ||
| 4 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Graduate School of Life Science and Systems En... | ur.014730512667.72 | Yusuke | Kamata | ||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 12176 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | 九州工業大学 | ur.012307631725.34 | 熊本 | 拓哉 | ||
| 12202 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Department of Bioscience and Bioinformatics, K... | ur.010340773642.58 | Satoshi | Fujii | ||
| 12706 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | 九州工業大学大学院情報工学研究院 | ur.010035501405.12 | 國近 | 秀信 | ||
| 12728 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | Graduate School of Life Science and Systems En... | ur.01004062131.45 | Momoko | Kumemura | ||
| 12752 | Kitakyushu | 1859307.0 | Japan | JP | grid.258806.1 | Kyushu Institute of Technology | 九工大 | ur.010024355257.03 | 河野 | 晴彦 |
740 rows × 12 columns
Also here, the procedure is exactly the same as in approach 1.
[17]:
authors_current_v2 = pubs_v2.as_dataframe_authors()
authors_current_v2 = authors_current_v2[authors_current_v2['current_organization_id'] == GRIDID].copy()
authors_current_v2.drop(columns=['pub_id'], inplace=True)
authors_current_v2.drop_duplicates('researcher_id', inplace=True)
print(f"===\nResearchers with affiliation to {GRIDID} at the time of speaking:", authors_current_v2.researcher_id.nunique(), "\n===")
authors_current_v2
===
Researchers with affiliation to grid.258806.1 at the time of speaking: 584
===
[17]:
| affiliations | corresponding | current_organization_id | first_name | last_name | orcid | raw_affiliation | researcher_id | |
|---|---|---|---|---|---|---|---|---|
| 0 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Siowhwa | Teo | [0000-0001-9927-8309] | [Graduate School of Life Science and Systems E... | ur.010525770330.49 | |
| 2 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Zhenhua | Xu | None | [Graduate School of Life Science and Systems E... | ur.012076416030.50 | |
| 3 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Chu | Zhang | None | [Graduate School of Life Science and Systems E... | ur.010227671107.36 | |
| 4 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Yusuke | Kamata | None | [Graduate School of Life Science and Systems E... | ur.014730512667.72 | |
| 6 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Tingli | Ma | [0000-0002-3310-459X] | [Graduate School of Life Science and Systems E... | ur.01144540527.52 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 10482 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | 毛利 | 恵美子 | None | [九州工業大学大学院] | ur.010435415335.62 | |
| 10525 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | 田中 | 和博 | None | [九工大] | ur.010456027505.69 | |
| 11311 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | 國近 | 秀信 | None | [九州工業大学大学院情報工学研究院] | ur.010035501405.12 | |
| 11328 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | Momoko | Kumemura | None | [Graduate School of Life Science and Systems E... | ur.01004062131.45 | |
| 11344 | [{'city': 'Kitakyushu', 'city_id': 1859307, 'c... | grid.258806.1 | 河野 | 晴彦 | None | [九工大] | ur.010024355257.03 |
584 rows × 8 columns
Conclusions¶
As anticipated above, both approaches are equally valid and in fact they return the same (or very similar) number of results. Let’s compare them:
[18]:
# create summary table
data = [['1', len(authors_current), len(authors_historical),
"""search publications where year in [2013:2018] and research_orgs="grid.258806.1" return publication""",
],
['2', len(authors_current_v2), len(authors_historical_v2),
"""search researchers where research_orgs="grid.258806.1" return researchers --- then --- search publications where researchers.id in {IDS} and year in [2013:2018] and research_orgs={GRIDID} return publications""",
]]
pd.DataFrame(data, columns = ['Method', 'Authors (current)', 'Authors (historical)', 'Query'])
[18]:
| Method | Authors (current) | Authors (historical) | Query | |
|---|---|---|---|---|
| 0 | 1 | 584 | 830 | search publications where year in [2013:2018] ... |
| 1 | 2 | 584 | 740 | search researchers where research_orgs="grid.2... |
In some cases you might encounter small differences in the total number of records returned by the two approaches (eg one method returns 1-2 extra records than the other one).
This is usually due to a synchronization delay between Dimensions databases (e.g. publications and researchers). The differences are negligible in most cases, but in general it’s enough to run same extraction again after a day or two for the problem to disappear.
It depends on which/how many filters are being used in order to identify a suitable results set for your research question.
The first approach is generally quicker as it has only two steps, as opposed to the second method that has three.
However if your initial
publicationsquery returns lots of results (eg for a large institution or big time frame), it may be quicker to try out method 2 instead.The second approach can be handy if one wants to pre-filter researchers using one of the ther available properties (e.g.
last_grant_year)
So, in general, deciding which approach is best depends on the specific query filters being used and on the impact these filters have on the overall performance/speed of the data extraction.
There is no fixed rule and a bit of trial & error can go a long way in helping you optimize your data extraction algorithm!
Note
The Dimensions Analytics API allows to carry out sophisticated research data analytics tasks like the ones described on this website. Check out also the associated Github repository for examples, the source code of these tutorials and much more.